Inaugural Krantz Awards Recipient
2023 Breakthrough Award: Exploring cancer metabolism to direct treatments
Team: Nabeel Bardeesy, PhD and Raul Mostoslavsky, MD, PhD.
Learn more about the team's project and the Krantz Awards
Research Summary
Pancreatic cancer and biliary cancer are among the most lethal types of human cancers. The Bardeesy laboratory has developed a series of genetically engineered mouse models and patient-derived models to define the role of key gene mutations that drive these cancer types. Current projects focus on understanding the function of cancer genes in controlling the way cells modulate their growth and utilize energy in response to available nutrients. Additional studies are exploring how some therapies targeting key mutations initially cause tumor to stop growth and why resistance eventually develops. Each of these studies is being used to inform improved therapeutic approaches.
Research Projects
The Bardeesy lab studies the pathways driving the pathogenesis of pancreatic and biliary cancers. The lab has developed a series of genetically engineered mouse models that has elucidated the functional interactions of major gene mutations associated with these diseases in humans. Studies have focused on the roles of key cancer genes in regulation of cell metabolism, and the discovery of mechanisms of resistance to targeted therapies.
Interplay between metabolism and chromatin regulation
An important area of current focus in our lab is to elucidate the metabolic regulators of pancreatic cancer and biliary cancers, with particular attention paid to factors that reprogram cancer cell metabolism. We have linked mutations in the IDH1 gene to changes in metabolism that ultimately alter epigenetic states. Identifying these pathways has provided insights in mechanisms of cell transformation arising from these mutations and predict novel therapeutic vulnerabilities. Mutant IDH proteins acquire a novel enzymatic activity allowing them to convert alpha-ketoglutarate (αKG) to 2-hydroxyglutarate (2HG), which inhibits the activity of multiple αKGdependent dioxygenases, including the TET family DNA demethylases. We are focusing on how epigenetic defects caused by IDH-mediated inhibition of TET affect cross-talk between tumor and immune cells to support cancer growth.
Oncogenic functions of protein kinase A signaling in pancreatic and liver cancers
The protein kinase A (PKA) signaling pathway is activated by mutations in a number of tumor types. These include the subset of pancreatic and biliary tumors harboring mutations in GNAS, an upstream regulator of PKA, and a type of liver tumor (fibrolamellar carcinoma) harboring activating gene fusions of PKA. Although PKA is an important driver of the growth of these tumor types, the specific oncogenic mechanisms have not been as widely studied as for many other cancer gene mutations. We have focused on elucidating the primary mechanisms of PKA-driven growth. Our work has identified the Salt-inducible kinases (SIK1-3) as the critical targets of cancer-causing PKA alterations. In addition, we have linked this pathway to a downstream epigenetic mechanism controlling proliferation and reprogramming mitochondrial function and tumor cell metabolism.
Understanding and targeting FGFR2-driven biliary cancer
Genetic alterations that activate Fibroblast Growth Factor 2 (FGFR2) signaling are common in biliary cancer and predict response to pharmacological inhibition of the FGFR in patients. However, tumor shrinkage is often modest and acquired resistance invariably arises. We are investigating oncogenic mechanisms controlled by FGFR2 in biliary cancer, including direct targets of FGFR2 signaling as well as downstream impact on cellular metabolism and differentiation. Additionally, we are investigating resistance mechanisms and approaches to prevent and overcome resistance.
Models of biliary cancer
Recent genetic studies have identified multiple recurrent mutations in biliary cancers and have indicated considerable genetic heterogeneity between individual tumors. A key limitation in the field includes a paucity of experimental systems with which to define the contributions of the lesions to biliary cancer progression. We have established a series of genetically engineered mouse models that incorporate combinations of the major mutations found in the human disease. In addition, our ongoing efforts include the development of a human biliary cancer cell line bank and the use of this system in large-scale genetic and small-molecule screens to systematically define targetable vulnerabilities in molecularly defined subtypes of this cancer.
Research Positions
Postdoctoral Position - Bardeesy Lab
A Postdoctoral Research Fellow position is available to study molecular pathways in the pathogenesis of pancreatic and liver cancer, focusing regulators of cell metabolism and epigenetics. The candidate is expected to have a PhD in the biological sciences, and be highly motivated with expertise in basic molecular biology and biochemical techniques. The Fellow will have simultaneous academic appointments at the Massachusetts General Hospital and Harvard Medical School. Studies will involve the use of a number of genetic and biochemical approaches, including genetically engineered mouse models, primary epithelial cell systems, genome-wide analyses, and in vitro genetic screens to study the interplay of epigenetics and metabolism governing cancer initiation and progression. The position provides a rich intellectual environment within a group of young investigators at the MGH Cancer Center and Center for Regenerative Medicine, with full integration into the large research communities of the Massachusetts General Hospital and Harvard.
To apply, please email a brief cover letter and CV to nelbardeesy@partners.org.
Publications
Selected Publications
Gritti I, Wan J, Weerasekara V, Vaz JM, Tarantino G, Bryde TH, Vijay V, Kammula AV, Kattel P, Zhu S, Vu P, Chan M, Wu MJ, Gordan JD, Patra KC, Silveira VS, Manguso RT, Wein MN, Ott CJ, Qi J, Liu D, Sakamoto K, Gujral TS, Bardeesy N. DNAJB1-PRKACA fusion drives fibrolamellar liver cancer through impaired SIK signaling and CRTC2/p300-mediated transcriptional reprogramming. Cancer Discovery 2024 (in press).
Wu MJ, Kondo H, Kammula AV, Shi L, Xiao Y, Dhiab S, Xu Q, Slater CJ, Avila OI, Merritt J, Kato H, Kattel P, Sussman J, Gritti I, Eccleston J, Sun Y, Cho HM, Olander K, Katsuda T, Shi DD, Savani MR, Smith BC, Cleary JM, Mostoslavsky R, Vijay V, Kitagawa Y, Wakimoto H, Jenkins RW, Yates KB, Paik J, Tassinari A, Saatcioglu DH, Tron AE, Haas W, Cahill D, McBrayer SK, Manguso RT, Bardeesy N. Mutant IDH1 inhibition induces dsDNA sensing to activate tumor immunity. Science. 2024 Jul 12;385(6705)
Zhen Y, Liu K, Shi L, Shah S, Xu Q, Ellis H, Balasooriya ER, Kreuzer J, Morris R, Baldwin AS, Juric D, Haas W, Bardeesy N. FGFR inhibition blocks NF-ĸBdependent glucose metabolism and confers metabolic vulnerabilities in cholangiocarcinoma. Nature Communications 2024 May 30;15(1):4099.
Shi L, Shen W, Davis MI, Kong K, Vu P, Saha SK, Adil R, Kreuzer J, Egan R, Lee TD, Greninger P, Shrimp JH, Zhao W, Wei TY, Zhou M, Eccleston J, Sussman J, Manocha U, Weerasekara V, Kondo H, Vijay V, Wu MJ, Kearney SE, Ho J, McClanaghan J, Murchie E, Crowther GS, Patnaik S, Boxer MB, Shen M, Ting DT, Kim WY, Stanger BZ, Deshpande V, Ferrone CR, Benes CH, Haas W, Hall MD, Bardeesy N. SULT1A1- dependent sulfonation of alkylators is a lineage-dependent vulnerability of liver cancers. Nat Cancer. 2023 Mar;4(3):365-381.
Wu MJ, Shi L, Dubrot J, Merritt J, Vijay V, Wei TY, Kessler E, Olander KE, Adil R, Pankaj A, Tummala KS,..., Saad-Berreta R, Jenkins RW, Wang T, Heikenwälder M, Ferrone CR, Goyal L, Nicolay B, Deshpande V, Kohli RM, Zheng H, Manguso RT, Bardeesy N. Mutant IDH Inhibits IFNγ-TET2 Signaling to Promote Immunoevasion and Tumor Maintenance in Cholangiocarcinoma. Cancer Discov. 2022 Mar 1; 12(3):812-835.
Wu, Q, Zhen, Y, Shi, L, Vu P, Greninger P, Adil R, Merritt J, Egan R, Wu MJ, Yin X, Ferrone CR, Deshpande V, Baiev I, Pinto CJ, McLoughlin DE, Walmsley CS, Stone JR, Gordan JD, Zhu AX, Juric D, Goyal L, Benes CH, Bardeesy N.. EGFR inhibition potentiates FGFR inhibitor therapy and overcomes resistance in FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov. 2022 May 2;12(5):1378-1395.
*Co-corresponding authors



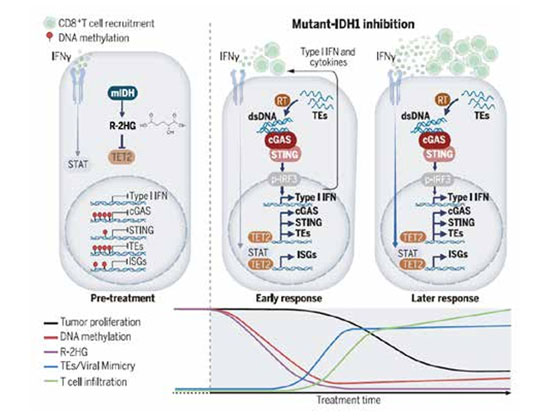 Cancers with mutant IDH1 (mIDH1) accumulate high levels of the oncometabolite R-2HG, which inhibits the DNA-demethylating enzyme TET2, leading to defects in immune signaling pathways. Pharmacological inhibition of mIDH1 activates TET2, which induces dormant virallike TEs throughout the genome and reactivates silenced genes in the cGAS-STING pathway that detect dsDNA generated by the TE-encoded reverse transcriptase. TET2 activation also enables tumor cells to respond to the antitumor cytokine IFNg. These immune pathways cooperate to promote CD8+ T cell infiltration and elicit a robust immune response.
Cancers with mutant IDH1 (mIDH1) accumulate high levels of the oncometabolite R-2HG, which inhibits the DNA-demethylating enzyme TET2, leading to defects in immune signaling pathways. Pharmacological inhibition of mIDH1 activates TET2, which induces dormant virallike TEs throughout the genome and reactivates silenced genes in the cGAS-STING pathway that detect dsDNA generated by the TE-encoded reverse transcriptase. TET2 activation also enables tumor cells to respond to the antitumor cytokine IFNg. These immune pathways cooperate to promote CD8+ T cell infiltration and elicit a robust immune response.