2024 Krantz Awards Recipient
2024 Quantum Award: Boosting immunotherapy response while eradicating toxicity
Team: Lloyd Bod, PhD, William L. Hwang, MD, PhD, David T. Ting, MD, and Alexandra-Chloe Villani, PhD.
Learn about the team's project and the Krantz Awards
Overview
The Villani laboratory seeks to establish a comprehensive roadmap of the human immune system by achieving a higher resolution definition and functional characterization of cell subsets and rules governing immune response regulation, as a foundation to decipher how immunity is dysregulated in diseases. We use unbiased systems immunology approaches, cutting-edge immunogenomics, single-cell ‘multi-omics’ strategies, and integrative computational frameworks to empower the study and modeling of the immune system as a function of “healthy” and inflammatory states, disease progression, and response to treatment. Our multi-disciplinary team of immunologists, geneticist, computational biologists, and physicians work towards answering several key questions:
- Do we know all existing blood immune cell subsets?
- How do circulating immune cells mirror those in tissue microenvironment in the context of health and disease?
- Can we identify targets that would improve immunotherapy efficacy by increasing specificity?
Collectively, our groundwork is paving the way for developing a human immune lexicon that is key to promoting effective bench-to-beside translation of findings.
Research Projects
Leveraging single-cell ‘multi-omics’ to unravel new insights into the human immune system
Achieving detailed understanding of the composition and function of the immune system at the fundamental unit of life — the cell — is essential to determining the prerequisites of health and disease. Historically, leukocyte populations have been defined by a combination of morphology, localization, functions, developmental origins, and the expression of a restricted set of markers. These strategies are inherently biased and recognized today as inadequate. Single-cell RNA sequencing (scRNAseq) and ‘multi-omics’ analysis provides an unbiased, data-driven way of systematically detecting cellular states that can reveal diverse simultaneous facets of cellular identity, from discrete cell types to continuous dynamic transitions, which cannot be defined by a handful of pre-defined markers or for which markers are not yet known. We combine scRNAseq strategies together with in-depth follow-up profiling, phenotypic and functional characterization of prospectively isolated immune subsets defined by scRNAseq data to overcome such limitations. Our analyses of the human blood mononuclear phagocyte system resulted in the identification of six dendritic cell (DC), four monocyte, and one DC progenitor populations, thus revising the taxonomy of these cells (Villani et al., Science 2017). Noteworthy, five of these subsets had never been reported, illustrating the power of our integrative strategies to reopen the definition of these cell types. Our study highlighted the value of embarking on a comprehensive Human Cell Atlas initiative and offered a useful framework for conducting this kind of analysis on other cell types and tissues. We are currently contributing to the immune cell atlas effort by charting at high-resolution the human blood cellular landscape, and are studying paired human tissues with blood to better establish how circulating immune cells mirror those in tissue microenvironment in the context of health and disease.
We also continuously support development of in-depth expertise in single-cell ‘multiomics’ experimental and computational strategies (Fisher F, Nat Commun 2024; Ding, Nat Biotechnol 2020; Li, Nat Methods 2020; Tukiainen, Villani, Nature 2017; Ranu, Villani, Nucleic Acid Res 2019; Villani, Methods Mol Biol 2016), and its application to study immune cells infiltrates in healthy, tumor lesions and inflamed tissue (Izar, Science 2016; Sade-Feldman, Cell 2019; Di Pilato, Nature 2019; Olah, Nat Commun 2018; Balan, Cell Rep 2018; Popescu, Nature 2019; Smillie, Cell 2019; Abbas, Nat Immunol 2020; Delorey, Nature 2021; Alladina, Science Immunol 2023).
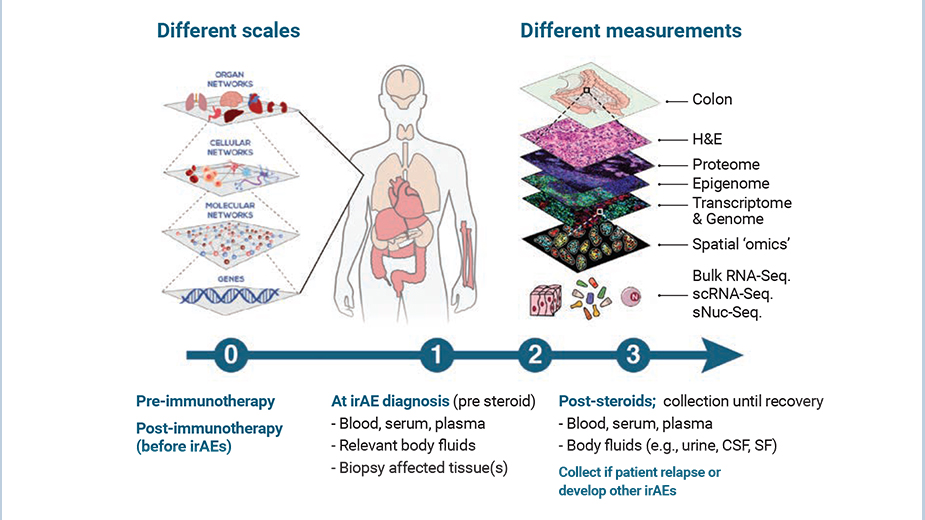
Deciphering immune-related adverse events (irAEs) induced by immunecheckpoint inhibitor (ICI) therapy
While ICI therapy is revolutionizing the treatment of solid cancers, its success is currently being limited by treatment-induced irAEs resembling autoimmune diseases that are affecting nearly every organ system. With ICI becoming first- and second-line of cancer treatments, it is expected that irAE incidence will continue rising and limit immunotherapy efficacy unless we find solutions. Our multi-disciplinary translational group of scientists and clinicians are working towards developing a better understanding of the biological players and underlying molecular and cellular mechanisms involved in driving irAEs by directly studying patient blood and matched affected tissue samples using a range of systems immunology, immunogenomics and single-cell ‘omics’ strategies (Zubiri, J Immunother Cancer 2021; Thomas, Nature Med 2024; Blum, Nature 2024). Our translational research program may result in identifying putative cellular components and mechanisms that could be (i) targeted in a ‘primary-prevention’ approach to prevent irAE development, and/or (ii) targeted after onset of irAEs, without reducing the efficacy of the immunotherapy.
Publications
Click here to view all publications from this investigator.
Selected Publications
Thomas MF*, Slowikowski K*, Mana- kongtreecheep K, Pritha Sen, …, Li B, Reynolds KL†, Villani AC†. Altered interactions between circulating and tissue- resident CD8 T cells with the colonic mucosa define colitis associated with immune checkpoint inhibitors. Nature Medicine 2024; 30 (5): 1349-1362 .
Alladina J*, Smith NP*, Kooistra T, Slowikowski K, Kernin IJ, Deguine J, Keen HL, Manakongtreecheep K, Tantivit J, Rahimi RA, Sheng SL, Nguyen ND, Haring AM, Giacona FL, Hariri LP, Xavier RJ, Luster AD, Villani AC†, Cho JL†, Medoff BD†. A human model of asthma exacerbation reveals transcriptional programs and cell circuits specific to allergic asthma. Sci Immunol. 2023; 8(83): eabq6352.
Zubiri L, Molina GE, Mooradian MJ, Cohen J, Durbin SM, Petrillo L, Boland GM, Juric D, Dougan M, Thomas MF, Faje AT, Rengarajan M, Guidon AC, Chen ST, Okin D, Medoff BD, Nasrallah M, Kohler MJ, Schoenfeld SR, Karp-Leaf RS, Sise ME, Neilan TG, Zlotoff DA, Farmer JR, Bardia A, Sullivan RJ, Blum SM, Semenov YR, Villani AC†, Reynolds KL†. Effect of a multidisciplinary Severe Immunotherapy Complications Service on outcomes for patients receiving immune checkpoint inhibitor therapy for cancer. J Immunother Cancer. 2021; 9(9): e002886.
Delorey TM*, Ziegler CGK*, Heimberg G*, Normand R*, Yang Y, Segerstolpe A, Abbondanza D, Fleming SJ, Subramanian A, Montoro DT, Jagadeesh KA, Dey KK, Sen P, Slyper M, Pita-Juárez YH, Phillips D, Biermann J, Bloom-Ackermann Z, Barkas N, Ganna A, Gomez J, Melms JC, Katsyv I, Normandin E, Naderi P, Popov YV, Raju SS, Niezen S, Tsai LT-Y, Siddle KJ, Sud M, Tran VM, Vellarikkal SK, Wang Y, Amir-Zilberstein L, Atri DS, Beechem J, Brook OR, Chen J, Divakar P, Dorceus P, Engreitz JM, Essene A, Fitzgerald DM, Fropf R, Gazal S, Gould J, Grzyb J, Harvey T, Hecht J, Hether T, Jané-Valbuena J, Leney-Greene M, Ma H, McCabe C, McLoughlin DE, Miller EM, Muus C, Niemi M, Padera R, Pan L, Pant D, Pe'er C, Pfiffner-Borges J, Pinto CJ, Plaisted J, Reeves J, Ross M, Rudy M, Rueckert EH, Siciliano M, Sturm A, Todres E, Waghray A, Warren S, Zhang S, Zollinger DR, Cosimi L, Gupta RM, Hacohen N, Hibshoosh H, Hide W, Price AL, Rajagopal J, Tata PR, Riedel S, Szabo G, Tickle TL, Ellinor PT, Hung D, Sabeti PC, Novak R, Rogers R, Ingber DE, Jiang ZG, Juric D, Babadi M, Farhi SL, Izar B, Stone JR, Vlachos IS, Solomon IH, Ashenberg O, Porter CBM, Li B, Shalek AK†, Villani AC†, Rozenblatt-Rosen O†, Regev A†. . COVID-19 tissue atlases reveal SARS- CoV-2 pathology and cellular targets. Nature 2021; 595(7865):107-113. PMID: 33915569.
Villani AC*, Satija R*, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, Dixon D, Stephenson E, Nilsson E, Grundberg I, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A†, Hacohen N†. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes and progenitors. Science 2017; 356: 6335. pii: eaah4573.
*Co-first authorship
†Co-senior authorship