Goldberg Laboratory: Marcia B. Goldberg, MD
Contact Information
Explore This Laboratory
Our Research
Our research focuses on two related scientific ideas:
- The molecular nature of interactions between microbial pathogens and the host, with an emphasis on the intracellular human pathogen Shigella, a major cause of diarrhea and dysentery
- The development of rapid diagnostics for infectious diseases.
Research Projects
Alteration of host cell biology by bacterial pathogens
Our research focuses on the interface of bacterial pathogens with human cells. Most gram-negative bacterial pathogens use specialized secretion systems (“type III secretion systems”) to deliver virulence proteins into human cells during infection. The proteins that are delivered by this means (“effector proteins”) manipulate human signaling pathways, cytoskeletal dynamics, and innate and adaptive immune responses, in ways that promote disease. These effector proteins are absolutely necessary for disease. Using genome-wide screens, we identified human signaling pathways required for infection by the gram-negative bacterial pathogen Shigella flexneri. Shigella are important human pathogens that cause diarrhea and dysentery, predominantly in daycare centers, institutions, and developing regions of the world.
We investigate:
- The molecular mechanisms by which bacterial effector proteins alter cellular targets and cellular signaling pathways
- The mechanisms by which human signaling pathways identified in our screens contribute to infection
Images
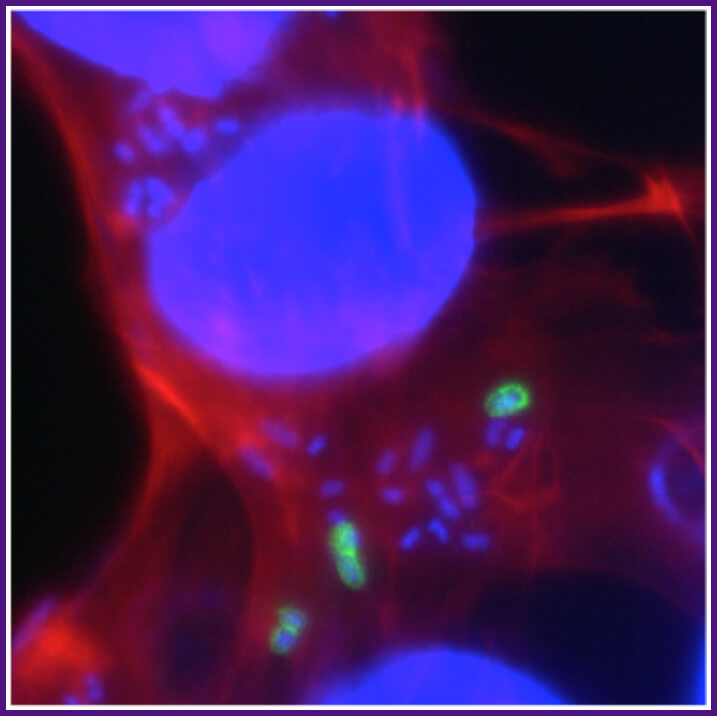 Shigella triggers its own entry into intestinal epithelial cells. Uptake of Shigella occurs upon activation of host cell actin polymerization and the formation of plasma membrane extensions that engulf the bacteria. The Shigella proteins that cause actin polymerization and membrane extensions are delivered into the cell via a specialized secretion system, the type 3 secretion system. Red, phalloidin staining of cellular actin; blue, DAPI staining of bacterial and cellular DNA; green, bacteria that have not yet entered the cell.
Shigella triggers its own entry into intestinal epithelial cells. Uptake of Shigella occurs upon activation of host cell actin polymerization and the formation of plasma membrane extensions that engulf the bacteria. The Shigella proteins that cause actin polymerization and membrane extensions are delivered into the cell via a specialized secretion system, the type 3 secretion system. Red, phalloidin staining of cellular actin; blue, DAPI staining of bacterial and cellular DNA; green, bacteria that have not yet entered the cell.
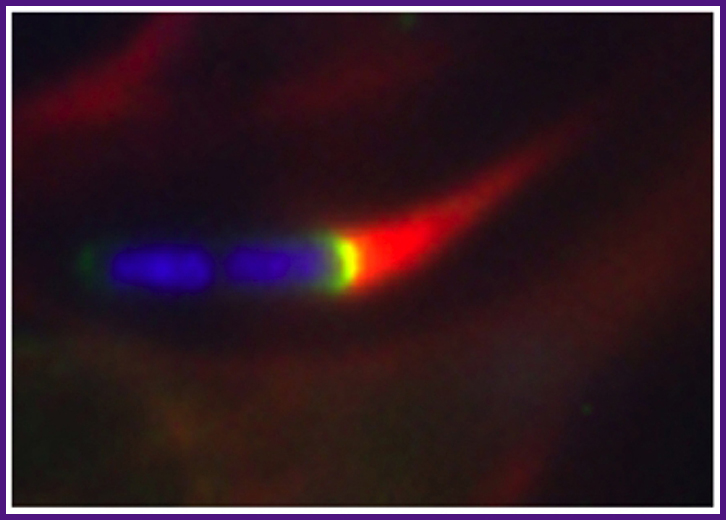 Shigella is able to move within and spread between cells by assembling actin at the pole of the bacterium, a process that generates force. In this fluorescence microscopy image, the bacterium is indicated in blue (DAPI staining of DNA), the comet-like actin tail in red (phalloidin staining of actin), and N-WASP, a cellular actin nucleation promoting factor recruited and activated by Shigella, in green (GFP-N-WASP).
Shigella is able to move within and spread between cells by assembling actin at the pole of the bacterium, a process that generates force. In this fluorescence microscopy image, the bacterium is indicated in blue (DAPI staining of DNA), the comet-like actin tail in red (phalloidin staining of actin), and N-WASP, a cellular actin nucleation promoting factor recruited and activated by Shigella, in green (GFP-N-WASP).
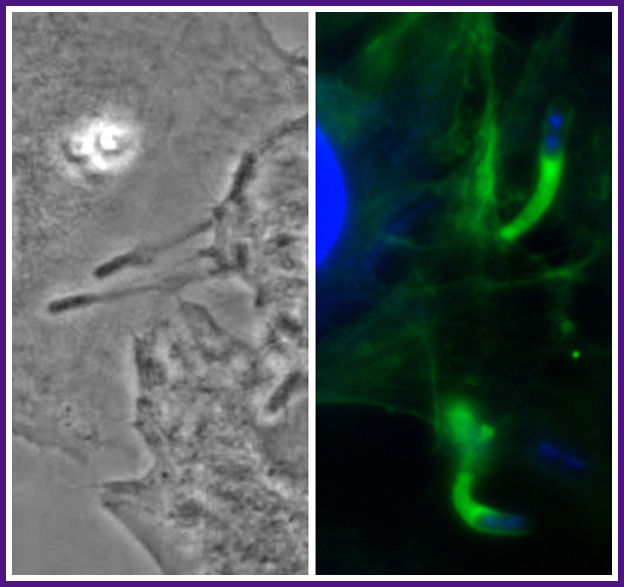 During Shigella spread from one cell into an adjacent cell, the bacterium pushes out within a sleeve of plasma membrane ("protrusion") that is subsequently engulfed by the adjacent cell. Left, phase micrograph showing Shigella at the tips of two protrusions. Right, fluorescent micrograph of Shigella at the tips of two protrusions. Green, tail of polymerized actin behind each Shigella bacterium; blue, DAPI staining of bacterial and cellular DNA.
During Shigella spread from one cell into an adjacent cell, the bacterium pushes out within a sleeve of plasma membrane ("protrusion") that is subsequently engulfed by the adjacent cell. Left, phase micrograph showing Shigella at the tips of two protrusions. Right, fluorescent micrograph of Shigella at the tips of two protrusions. Green, tail of polymerized actin behind each Shigella bacterium; blue, DAPI staining of bacterial and cellular DNA.
 Like many other gram-negative bacterial pathogens, Shigella delivers proteins into the cytoplasm of host cells from outside the cell. The proteins that are delivered—effector proteins—alter the function of host proteins in ways that promote bacterial infection. The apparatus by which these proteins are delivered—the type 3 secretion apparatus—consists of a long needle-like structure that extends from the bacterial surface. When the tip of the needle contacts with the plasma membrane, two bacterial proteins are secreted through the needle that form a pore (translocon pore) in the plasma membrane. The needle docks onto the pore, thereby creating a continuous channel from the bacterial cytoplasm (bottom of image) to the host cell cytoplasm (top of image), through which effector proteins are delivered. We showed that host cell intermediate filaments bind one of the pore proteins and that this binding interaction is required for docking of the needle onto the pore (Russo BC et al., Nat Microbiol, 2016).
Like many other gram-negative bacterial pathogens, Shigella delivers proteins into the cytoplasm of host cells from outside the cell. The proteins that are delivered—effector proteins—alter the function of host proteins in ways that promote bacterial infection. The apparatus by which these proteins are delivered—the type 3 secretion apparatus—consists of a long needle-like structure that extends from the bacterial surface. When the tip of the needle contacts with the plasma membrane, two bacterial proteins are secreted through the needle that form a pore (translocon pore) in the plasma membrane. The needle docks onto the pore, thereby creating a continuous channel from the bacterial cytoplasm (bottom of image) to the host cell cytoplasm (top of image), through which effector proteins are delivered. We showed that host cell intermediate filaments bind one of the pore proteins and that this binding interaction is required for docking of the needle onto the pore (Russo BC et al., Nat Microbiol, 2016).
Publications
View a full list of our publications on PubMed.
Filbin MR, Mehta A, …., Hacohen N, Goldberg MB. Plasma proteomics reveals tissue-specific cell death and mediators of cell-cell interactions in severe COVID-19 patients. Cell Rep Med. https://doi.org/10.1101/2020.11.02.365536.
Meyerowitz EA, Sanchez S, Mansour MK, Triant VA, Goldberg MB. Isolated cerebral mucormycosis in immunocompetent adults who inject drugs: case reports and systemic review of the literature. Open Forum Infectious Diseases. 2020. 7(12), ofaa552.
Duncan-Lowey JK, Wiscovitch AL, Wood TE, Goldberg MB#, Russo BC#. Shigella flexneri disruption of cellular tension promotes intercellular spread. Cell Rep. 2020; 33(8):108409. #Co-corresponding authors.
Kutsch M, Sistemich L, Lesser CF, Goldberg MB, Herrmann C, Coers J. Direct binding of polymeric GBP1 to LPS disrupts bacterial cell envelope functions. EMBO J. 2020; 39(13):e104926. PMC7327485.
Shrock E, Fujimura,…Goldberg MB, et al. Viral epitope profiling in COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020 Nov 27; 370(6520):eabd4250. doi: 10.1126/science.abd4250.
Reyes M, Filbin MR, Bhattacharyya RP, Billman K, Eisenhaure T, Hung DT, Levy BD, Baron RM, Blainey PC#, Goldberg MB#, Hacohen N#. An immune cell signature of bacterial sepsis. Nat Med. 2020 26:333-340. #Co-corresponding authors.
Russo BC, Duncan JK, Wiscovitch AL, Hachey AC, Goldberg MB. Activation of Shigella flexneri type 3 secretion requires a host-induced conformational change to the translocon pore. PLoS Pathog. 2019; 15:e1007928.
Russo BC, Duncan JK, Goldberg MB. Topological analysis of the type 3 secretion system translocon pore protein IpaC following its native delivery to the plasma membrane during infection. mBio. 2019 May 28;10(3). pii: e00877-19. doi: 10.1128/mBio.00877-19. *Editor’s Pick.
Bhattacharyya RP, Walker M, Boykin R, Son S, Liu J, Hachey AC, Ma P, Wu L, Choi K, Cummins KC, Benson M, Skerry J, Ryu H, Wong SY, Goldberg MB, Han J, Pierce VM, Cosimi LA, Shoresh N, Livny J, Beechem J, Hung DT. Rapid identification and phylogenetic classification of diverse bacterial pathogens in a multiplexed hybridization assay targeting ribosomal RNA. Sci Rep. 2019; 9:4516.
Piantadosi A, Kanjilal S, Ganesh V, Khanna A, Hyle EP, Rosand J, Bold T, Metsky HC, Lemieux J, Leone MJ, Freimark L, Matranga CB, Adams G, McGrath G, Zamirpour S, Telford S 3rd, Rosenberg E, Cho T, Frosch MP, Goldberg MB, Mukerji SS, Sabeti PC. Rapid Detection of Powassan Virus in a Patient With Encephalitis by Metagenomic Sequencing. Clin Infect Dis. 2018;66:789-792. PMC5850433.
Miller KA, Garza-Mayers AC, Leung Y, Goldberg MB. Identification of interactions among host and bacterial proteins and evaluation of their role early during Shigella flexneri. Microbiology. 2018; 164:540-550. DOI: 10.1099
Piantadosi A, Mukerji SS, Chitneni P, Cho TA, Cosimi LA, Hung DT, Goldberg MB, Sabeti PC, Kuritzkes DR, Grad YH. Metagenomic sequencing of an echovirus 30 genome from cerebrospinal fluid of a patient with aseptic meningitis and orchitis. Open Forum Infectious Diseases. 2017. 4:ofx138. PMC5524216.
Russo BC, Stamm LM, Raaben M, Kim CM, Kahoud E, Robinson LR, Bose S, Queiroz AL, Herrera BB, Baxt LA, Mor-Vaknin N, Fu Y, Molina G, Markovitz DM, Whelan SP, Goldberg MB. Intermediate filaments enable pathogen docking to trigger type 3 effector translocation. Nat Microbiol. 2016. Article No. 16025; DOI: 10.1038.
Lu R, Herrera BB, Eshleman HD, Fu Y, Bloom A, Li Z, Sacks DB, Goldberg MB. Shigella effector OspB activates mTORC1 in a manner that depends on IQGAP1 and promotes cell proliferation. PLoS Pathogens. 2015; 11(10):e1005200. PMC4608727.
Lee SY, Gertler FB, Goldberg MB. Vasodilator-stimulated phosphoprotein (VASP) restricts cell-to-cell spread of Shigella flexneri at the cell periphery. Microbiology. 2015; 161:2149-2160. PMC4806587.
Garza-Mayers AC, Miller KA, Russo BC, Nagda DV, Goldberg MB. Shigella flexneri regulation of ARF6 activation during bacterial entry via an IpgD-mediated positive feedback loop. mBio. 2015; 6(2). pii: e02584-14. PMC4358011
Yi C-R, Allen JE, Russo B, Lee SY, Heindl JE, Baxt LA, Herrera BB, Kahoud E, MacBeath G, Goldberg MB. Systematic analysis of bacterial effector-postsynaptic density 95/discs large/zonula occludens-1 (PDZ) interactions demonstrates Shigella OspE promotes PKC activation via PDLIM proteins. J Biol Chem. 2014; 289:30101-30113. PMC4208017
Baxt LA, Goldberg MB. Host and bacterial proteins that repress recruitment of LC3 to Shigella early during infection. PLoS One. 2014;9(4):e94653. PMC3983221
Li Z, Boyd D, Reindl M, Goldberg MB. Identification of YidC residues that define interactions with the Sec Apparatus. J Bacteriol. 2014;196(2):367-77. PMC3911256
Gray AN, Li Z, Henderson-Frost J, Goldberg MB. Biogenesis of YidC cytoplasmic membrane substrates is required for positioning of autotransporter IcsA at future poles. J Bacteriol. 2014;196(3):624-32. PMC3911155
Baxt LA, Garza-Mayers AC, Goldberg MB. Bacterial subversion of host innate immune pathways. Science. 2013;340(6133):697-701.
Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, Goldberg MB, Xavier RJ. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12(6):778-90. PMCID: PMC3785244
Fixen KR, Janakiraman A, Garrity S, Slade DJ, Gray AN, Karahan N, Hochschild A, Goldberg MB. Genetic reporter system for positioning of proteins at the bacterial pole. MBio. 2012;3(2). PMC3302567
Gray AN, Henderson-Frost JM, Boyd D, Shirafi S, Niki H, Goldberg MB. Unbalanced charge distribution as a determinant for dependence of a subset of E. coli membrane proteins on the membrane insertase YidC. MBio. 2011; 2(6): doi:10.1128/mBio.00238-11. PMC3911155
Stamm LM, Goldberg MB. Establishing the secretion hierarchy. Science. 2011 4;331(6021): 1147-1148.
Jehl S, Doling A, Giddings, KS, Phalipon A, Sansonetti PJ, Goldberg MB, Starnbach MN. Antigen-specific CD8+ T cells fail to respond to Shigella flexneri. Infect Immun. 2011: 79(5); 2021-2030. PMC3088127
Stamm LM, Heller DM, Goldberg MB. Caging targets for destruction. Cell Host Microbe. 2010: 8: 391-393.
Baxt LA, Goldberg MB. Anaerobic environment of the intestine primes pathogenic Shigella for infection. Expert Rev Anti Infect Ther. 2010: 8(11): 1225-1229.
Lu R, Goldberg MB. Bacterial Exploitation of Host Cell Signaling. Sci Transl Med. 2010; 2(51): 48.
Heindl JE, Saran I, Yi CR, Lesser CF, Goldberg MB. Requirement for formin-induced actin polymerization during spread of Shigella. Infect Immun. 2009 Jan; 78(1):193-203. PMC2798232
Yi CR, Goldberg MB. Enterohemorrhagic Escherichia coli raises the I-BAR. Proc Natl Acad Sci USA. 2009; 106(16): 6431-2.
Yi CR, Goldberg MB. Putting enterohemorrhagic E. coli on a pedestal. Cell Host Microbe. 2009; 5(3): 215-7.
Janakiraman A, Fixen KR, Gray AN, Niki H, Goldberg MB. A genome-scale screen identifies a role for DnaK in chaperoning of polar autotransporters in Shigella. J Bacteriol. 2009; 191(20): 6300-6311. PMC2753027
Wagner JK, Heindl JE, Gray AN, Jain S, Goldberg MB. Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J Bacteriol. 2009 Feb;191(3):815-821. PMC2632083
Group Members
Meet our research team.
-
![]()
- Infectious Diseases
- Department of Medicine
Brian C. Russo, PhD, Postdoctoral Fellow
Amanda S. Zajac, PhD, Postdoctoral Fellow
Poyin Chen, PhD, Postdoctoral Fellow
Maricarmen Rojas-Lopez, PhD, Postdoctoral Fellow
Thomas E. Wood, PhD, Postdoctoral Fellow
Jeffrey K. Duncan, Research Technician
MyungSeo Yoon, Research Student - Harvard University
Vritti Kharbanda, Summer Student - Brandeis University
Christian Caliboso, Summer Student - Harvard University
Julia Flores, Summer Student - Johns Hopkins University
A Top Hospital in America
Mass General is recognized as a top hospital on the U.S. News Best Hospitals Honor Roll for 2024-2025.
Patient and Family Relations
Patient and Family Relations specialists can help patients and families resolve issues or express praise or concern about their experience at MGH.

