Krantz Family Center for Cancer Research
Vázquez-García Laboratory
Contact Information
Vázquez-García Laboratory
Ignacio Vazquez-Garcia, PhD
Assistant Professor of Pathology
Mass General Brigham Cancer Institute
Harvard Medical School
Program Affiliations
Krantz Family Center for Cancer Research
Explore the Vázquez-García Lab
Overview

The Vázquez-García Laboratory develops quantitative approaches to study somatic evolution in cancer, focusing on the rapidly evolving genomes of tumors and their interactions with the immune system. Throughout life, somatic cells acquire genetic and epigenetic alterations, forming a mosaic of mutant clones that typically remain benign. Genome instability disrupts this balance, driving malignant transformation and generating tumor clones with distinct evolutionary capacities that fuel immune evasion, metastasis, and therapy resistance. We integrate innovations in AI/ML and high-throughput genomics to dissect how genome instability shapes tumor evolution and immune dynamics. New single-cell and spatial technologies enable large-scale profiling of genomic, transcriptomic, and epigenetic states, revealing intra-tumoral heterogeneity, spatial organization, and temporal dynamics. Leveraging multimodal data with advanced computational models, we uncover how genomic adaptations confer selective advantages to tumor cells and how they activate, evade, or suppress immune responses. Our ultimate goal is to define the principles of somatic evolution and immune surveillance, and to translate these insights into predictive biomarkers and therapeutic strategies that anticipate and overcome tumor progression.
Research Summary
Single-cell cancer dynamics
Cancer evolves through the continual acquisition of somatic alterations, with genome instability acting as a catalyst for rapid diversification, clonal evolution and clinical progression. Large-scale genomic alterations such as whole-genome doubling, chromosomal copy number changes, and extrachromosomal DNA dominate the evolutionary landscape of many cancers. These mutational processes precipitate intra-tumor diversification and shape recurrent evolutionary trajectories across cancers. Our work leverages single-cell genomics and spatial technologies to resolve how these processes emerge, propagate, and influence tumor progression, therapeutic resistance, and metastatic potential. To advance these efforts, we have pioneered high-throughput single-cell genome sequencing technologies, applying them in high-grade serous ovarian cancer as an archetypal tumor driven by genome instability. These single-cell approaches can reveal genomic alterations arising from individual cell division errors and enable lineage tracing using native markers. Integrating these measurements with longitudinal monitoring and clinical data, we aim to define conserved evolutionary trajectories and mechanisms of genome rearrangement driving diversity, resistance, and metastasis.
Genetic alterations alone rarely explain why some mutant clones remain dormant while others progress, adapt, or metastasize. Genome instability drives widespread aneuploidy, altering gene dosage across hundreds of loci and disrupting cellular processes by rewiring transcriptional and epigenetic programs. These dosage-driven effects, combined with transcriptional and epigenetic plasticity, generate dynamic and reversible phenotypes that enable therapy resistance and metastatic dissemination. Using single-cell multi-omics, we are mapping genotype-phenotype relationships to identify the regulatory mechanisms that shape these adaptive states. We are also engineering defined genomic alterations and causally testing their impact on cellular fitness to reveal how genomic changes and phenotypic adaptability jointly promote tumor evolvability.
Tumor-immune co-evolution
Genome instability not only enhances tumor adaptation but also reshapes tumor-immune interactions, often driving immune suppression and resistance to immunotherapy. Emerging evidence suggests that genome instability can also activate immune signaling early in tumorigenesis, potentially triggering immune editing before tumors adapt to suppress these responses. To dissect these dynamics, we leverage single-cell and spatial technologies to map the determinants of immune recognition, avoidance, and evasion in genomically unstable cancers. In recent work, we found that distinct mutational processes shape ovarian cancer evolution by altering intra-tumoral immune phenotypes and enabling immune escape.
Building on these insights, we are developing predictive models that integrate tumor evolutionary history with spatially resolved phenotypes to capture the interplay between genome instability, antigen clonality, and immune selection. Unstable tumor genomes generate diverse immunogenic and tolerogenic signals. For example, chromosome missegregation and micronucleus rupture can release DNA into the cytosol and engage the innate immune system. At the same time, the clonal distribution of tumor-derived neoantigens modulates adaptive immune responses, influencing whether tumor clones are eliminated or persist. By mapping how somatic mutations accrue and spatially evolve under immune pressure, we aim to identify signatures distinguishing lesions that evade immune surveillance from those that elicit productive anti-tumor immunity. These insights will directly inform the design of immunotherapies to target genomically unstable clones that evade immune clearance.
Multimodal AI in cancer biology
To address these challenges, we develop multimodal AI frameworks that integrate genomic, transcriptomic, imaging, and clinical data into unified models of tumor evolution. We use representation learning as a foundational approach to align diverse single-cell and tissue-scale measurements into a shared latent space that captures the multi-scale organization of tumor ecosystems. Building on this, we leverage generative models, from single-cell dynamics to population-level frameworks, to simulate tumor and immune trajectories under therapeutic pressures. By contextualizing genomic variants within cellular states, embedding them in tissue architectures, and connecting these patterns to clinical phenotypes, we aim to move beyond static biomarkers toward dynamic, anticipatory frameworks that guide precision prevention and targeted intervention.
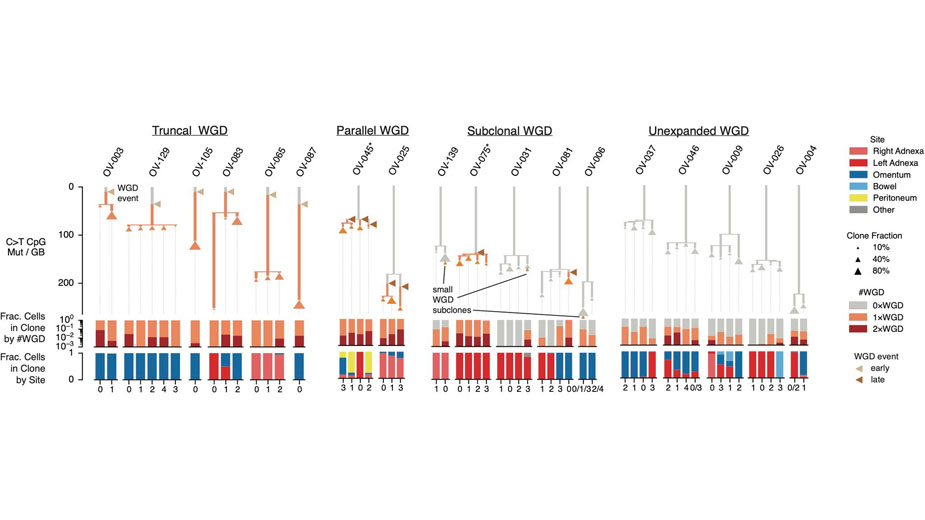
Research Image
Single-cell genome sequencing enables evolutionary timing of whole-genome doubling (WGD) events in ovarian cancer. The clone phylogenies illustrate the evolutionary histories and timing of WGD, revealing a complex role for WGD expansions across the evolutionary continuum. Branch length shows the number of age-associated SNVs (C-to-T at CpG sites) assigned to each branch, adjusted for coverage depth. Expanded WGD events are shown as triangles at the predicted location along WGD branches, colored by relative timing. Branches are colored by the number of WGD events in the evolutionary history of each tumor cell. Bar plots show, for each leaf, the fraction of cells grouped by the number of WGD events and the fraction of cells from each anatomical site.
Select Publications
McPherson A*, Vázquez-García I*, Myers M*, Al-Rawi D*, Zatzman M*, ..., Shah S#. Ongoing genome doubling shapes evolvability and immunity in ovarian cancer. Nature. 2025.
Williams M, Vázquez-García I, Tam G, Wu M, Varice N, …, Shah S#. Tracking clonal evolution during treatment in ovarian cancer using cfDNA. Nature. In press.
Dinh K#, Vázquez-García I#, Chan A, Malhotra R, Weiner A, McPherson A, and Tavaré S#. CINner: modeling and simulation of chromosomal instability in cancer at single-cell resolution. PLoS Comp Bio. 2025 21(4):e1012902.
Canadas I, Casirati G, Houlahan K, Maxwell K, Mehta A, Parolia A, Saulnier O, Taylor A, Thomas C, and Vázquez-García I. Insights on future directions in cancer research from the AACR NextGen Stars. Cancer Discovery. 2024 14(9):1584–1589.
Vázquez-García I*, Uhlitz F*, Ceglia N, Lim J, Wu M, …, Zamarin D#, Shah S#. Ovarian cancer mutational processes drive site-specific immune evasion. Nature. 2022 612:778–786.
Vázquez-García I#, Salinas F, Li J, Fischer A, Barré B, Hallin J, Bergström A, Alonso-Perez E, Warringer J, Mustonen V#, Liti G#. Clonal heterogeneity influences the fate of new adaptive mutations. Cell Reports. 2017 21(3):732-744.
*Equal contribution
#Corresponding authors
Lab Members
Oprah Nkera
Megan Tandar
Ignacio Vázquez-García, PhD
Jeffrey Wang
Xiwen Zhang
Krantz Family Center for Cancer Research
The scientific engine for discovery for the Mass General Brigham Cancer Institute.
Support the Krantz Family Center for Cancer Research
When you support us you are enabling discoveries that will lead to effective new weapons in the battle against cancer.
