Krantz Family Center for Cancer Research
Xia Laboratory
Contact Information
Xia Laboratory
Bo Xia, PhD
Assistant Professor of Pathology (Molecular Pathology Unit)
Mass General Brigham Cancer Institute
Harvard Medical School
Associate Member, Broad Institute of MIT and Harvard
Program Affiliations
Krantz Family Center for Cancer Research
Explore the Xia Lab
Overview

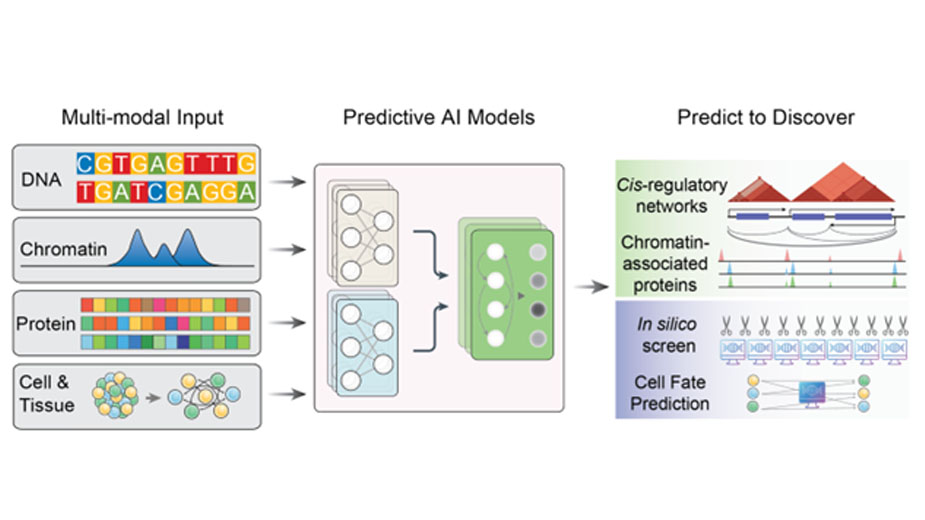
The Xia lab focuses on addressing one of biology’s most fundamental questions: how does the human genome control the identity and function of thousands of different cell types, and what goes wrong in diseases like cancer? To address this grand challenge, we need advanced technologies that exponentially accelerate the investigation pace beyond the traditional experimental approaches. In our lab, we develop state-of-the-art multimodal artificial intelligence (AI) tools that can predict how genes are regulated upon perturbations, allowing us to run large-scale in silico experiments before validating them in the lab. Using these technologies, we have discovered key proteins and genetic elements that shape how human genome folds and functions, and uncovered novel factors that regulate T cell fate transitions. By combining AI with advanced single-cell methods, our goal is to understand the core regulatory program that determine cell fate — and engineer these programs open new possibilities for regenerative medicine, aging research, and cancer therapy.
Research Summary
Multimodal AI Technology Development
How does the human genome encode the regulatory programs that specify thousands of cell types in health and disease? Genome regulation and cell fate determination are intrinsically multimodal, integrating DNA sequence, chromatin state, protein complexes, and their intricate interactions. My laboratory develops interdisciplinary technologies to address these challenges. Traditional experimental biology has relied on iterative cycles of observation, perturbation, and measurement—a process powerful in principle but inefficient at scale. To accelerate the investigation of cell fate determination, we build predictive AI/ML models and high-throughput in silico screening frameworks that extend the reach of experiments into computational space. Our team has developed foundational multimodal genomics AI models—including C.Origami (Nature Biotechnology, 2023) and Chromnitron (bioRxiv, 2025)—that capture the key principles of genome regulation and enable high-throughput computational genetic screens.
By coupling predictive genomics with targeted experimental validation, we aim to transform how fundamental discoveries are made, opening new avenues for manipulating cell fate and function. These technologies are designed not only to accelerate scientific progress but also to provide a generalizable platform with applications across cancer, regeneration, and aging.
Genome Regulation
The human genome encodes ~2,500 chromatin-associated proteins—including ~1,600 transcription factors—that collectively define the regulatory landscape. Understanding how DNA sequence, chromatin state, and protein networks specify gene expression remains one of the central unsolved problems in biology. The mammalian genome is hierarchically organized, with interactions spanning from local sequence motifs to higher-order 3D architecture. Dissecting this complexity requires new technologies.
We apply our multimodal genomics AI tools—trained on DNA sequence, accessibility, and protein features—to predict regulatory interactions and discover key regulators. By coupling these models with single-cell, spatial, and molecular assays, we investigate how gene regulation is orchestrated from base-pair resolution to chromosome-scale architecture. Recently, we identified ZNF654 and JMJD6 as long-sought regulators that work together with CTCF to shape mammalian genome organization (manuscript in preparation); and discovered novel factors that regulates T cell fate transition during chronic stimulation. We further extend these approaches to disease, examining how dysregulation of genome organization contributes to cancer and other pathologies, and how regulatory circuits can be manipulated, restored, or synthetically designed for therapeutic benefit.
Cellular Dynamics
At the cellular level, gene regulation underlies the extraordinary diversity and plasticity of mammalian cell types. Our work seeks to uncover the fundamental mechanisms by which regulatory programs drive cell fate transitions, tissue homeostasis, and organismal responses to stress and aging. We introduced the “periodic table of cell types” framework (Development, 2019), which organizes cellular identities across development. Using single-cell technologies, we previously charted spermatogenesis at unprecedented resolution (Cell, 2020) and mapped dynamic cellular interactions in the first pig-to-human kidney xenotransplantation (Med, 2024). More recently, we discovered the first plausible genetic mechanism that a single Alu element insertion in the ancestral genome may drive tail-loss evolution in humans and apes (Nature, 2024). This discovery also uncovered a previously unappreciated cis-regulatory mechanism that intronic repeat pairing can induce exon skipping splicing. Meanwhile, this discovery inspired a new investigation to control gene regulation as a new approach to chordoma, a rare tumor in spine or the base of skull.
Building on our earlier exploration of cellular dynamics in health and diseases, we now integrate predictive AI with gene regulatory analysis to address core questions: what factors ultimately determine a cell’s fate? How can we reprogram or de novo design cell types with novel functions? We focus particularly on chromatin-associated proteins, which form the regulatory circuits governing every cellular state. Through predictive modeling, single-cell epigenomics, and synthetic genetics, we aim to establish scalable approaches to engineer cell fate—providing insights for immunology, cancer biology, and aging biology.
Research Image
Select Publications
Tan, J.*, Fu, X.*, Ling, X.*, Mo, S., Bai, J., Rabadán, R., Fenyő, D., Boeke, J., Tsirigos, A#, Xia, B.# (2025) Multimodal learning decodes the global binding landscape of chromatin-associated proteins. BioRxiv. 2025.08.17.670761. [Link]
Xia, B.#, Zhang, W.*, Zhao, G.*, Zhang, X.*, Bai, J., Brosh, R., Wudzinska, A., Huang, E., Ashe, H., Ellis, G., Pour, M., Zhao, Y., Coelho, C., Zhu, Y., Miller, A., Dasen, J., Maurano, M., Kim, S., Boeke, J.#, and Yanai, I.# (2024) On the genetic basis of tail-loss evolution in humans and apes. Nature 626, 1042–1048. (Cover) [Link]
Pan, W.*, Zhang, W.*, Zheng, B.*, Camellato, B., Stern, J., Lin, Z., Khodadadi-Jamayran, A., Kim, J., Sommer, P., Khalil, K., Weldon, E., Bai, J., Zhu, Y., Meyn, P., Heguy, A., Mangiola, M., Griesemer, A., Keating, B.J.#, Montgomery, R.A.#, Xia, B.#, Boeke, J.D.#. Cellular dynamics of pig-to-human kidney xenotransplantation. Med. (2024), 5 (8), 1016-1029. (Cover) [Link]
Tan, J., Shenker-Tauris, N., Rodriguez-Hernaez, J., Wang, E., Sakellaropoulos, T., Boccalatte, F., Thandapani, P., Skok, J., Aifantis, I., Fenyo, D., Xia, B.#, Tsirigos, A.# (2023) Cell type-specific prediction of 3D chromatin organization enables high-throughput in silico genetic screening. Nature Biotechnology. 41(8), 1140-1150. [Link]
Xia, B., Yan, Y., Baron, M., Wagner, F., Barkley, D., Chiodin, M., Kim, S., Keefe, D., Alukal, J., Boeke, J. and Yanai, I. (2020) Widespread transcriptional scanning in the testis modulates gene evolution rates. Cell. 180 (2): 248-262. (Cover) [Link]
Xia, B.*, Han, D.*, Lu, X.*, Sun, Z., Zhou, A., Yin, Q., Zeng, H., Liu, M., Jiang, X., Xie, W., He, C.# and Yi, C.# (2015) Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nature Methods, 12(11): 1047–1050. [Link]
#corresponding author
*co-first authors
Lab Members
Bo Xia, PhD
Jiangshan 'Shawn' Bai (Research Associate)
Parmida Davarmanesh (Graduate Student)
Amulya Garimella (Rotation Student)
Xinyu Ling, PhD (Postdoc)
Qingji Lyu, PhD (Postdoc)
Jimin Tan, PhD (Visiting Scholar)
Krantz Family Center for Cancer Research
The scientific engine for discovery for the Mass General Brigham Cancer Institute.
Support the Krantz Family Center for Cancer Research
When you support us you are enabling discoveries that will lead to effective new weapons in the battle against cancer.
