Research Summary
The Hacohen laboratory consists of immunologists, geneticists, biochemists, technologists, physicians and computational biologists working together to develop new and unbiased technologies and strategies to understand basic immune processes and immune-mediated diseases, with an emphasis on the innate immunity, tool development and personalized medicine.
We address three key questions in immunology:
- How are immune responses against cancer initiated, maintained and evaded?
- What are the immune circuits that sense and control pathogens, such as viruses and bacteria?
- How does immunity against the body develop, in particular, in patients with autoimmune lupus?
In addition to discovering and studying specific molecular and cellular mechanisms, we also address how and why the immune response (to tumors, pathogens or self) varies so dramatically across individuals, such as in sepsis. Finally, we are adapting our unbiased analytical strategies into realworld therapeutics, having performed clinical trials (with our collaborator Dr. Catherine Wu), in which patients are vaccinated against their own tumors with a fully personal vaccine that is designed based on a computational analysis of their tumor genome.
Research Projects
Initiators, resistors and targets of tumor immunity
While cancer immunology has been deeply studied in animal models, there remain many open questions in human tumor immunology. We have developed genetic and genomics approaches to explain the large variance in anti-tumor immunity across people, and to discover how tumors evolve to resist productive immunity. We’ve identified somatic mutations in tumors that are associated with anti-tumor immunity in patients, found T cell subtypes that are associated with a response to anti- PD-1 immunotherapy in melanoma and are studying their properties now (Sade-Feldman et al., Cell 2018), and discovered spatiallyorganized immune cell hubs in colon cancer (Pelka, Hofree, Chen et al, Cell 2021; Chen et al, Nat Imm 2024). We have also developed new methods to predict which tumor antigens are presented (Abelin et al., Immunity 2017, Sarkizova et al., Nat Biotech 2020), which are now being used to develop novel therapeutic approaches and targets for immunotherapy, such as personal tumor vaccines targeting multiple HLA-associated neoantigens in human tumors (together with Dr. Catherine Wu at DFCI, Ott et al., Nature 2017, Keskin, Nature 2018).
Genes and networks underlying innate immunity
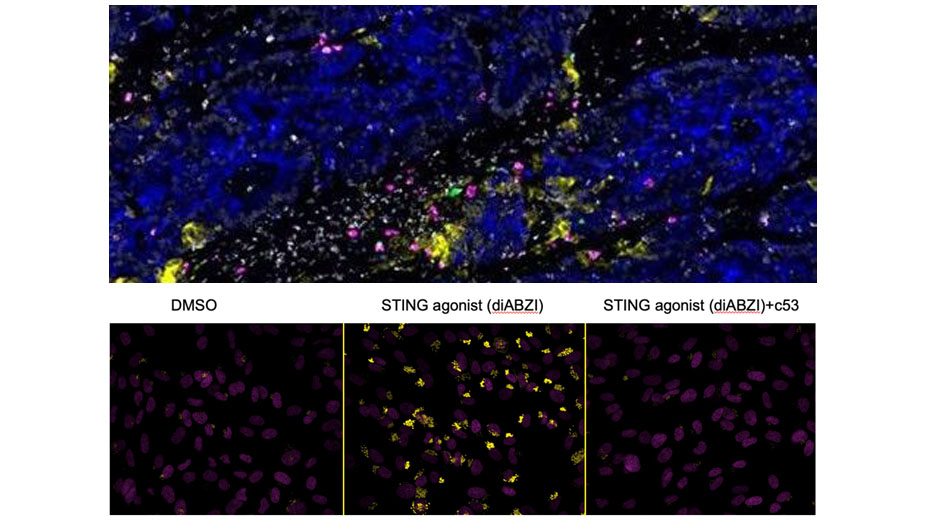
We’ve used genome-wide CRISPR libraries to discover mammalian genes mediating the sensing of pathogens (Parnas et al., Cell 2015), impacting HIV infection (Park et al, Nat Gen 2017) and affecting influenza infection (Li et al., Nat Comm 2020). We have characterized innate myeloid cells (DCs and monocytes) in human blood as part of the human Immune Cell Atlas (Villani et al, Science 2017). We defined regulators of viral RNA-sensing (Carlson et al., PNAS 2023) and DNA-sensing pathways using FACS- and imaging-based screens. Recently, we discovered that the STING protein, a protein required for sensing cyclic di-nucleotides, is a proton channel that can trigger LC3B lipidation, inflammasome activation and cell death (Liu, Carlson et al., Science 2023).
Genetic basis for inter-individual variations in immune responses
We have also developed genomic strategies to analyze human immune responses and explain immune phenotypes with germline genotypes. We characterized the genetic basis for inter-individual variation in the innate immune response to viruses and bacteria (Lee et al., Science 2014; Raj et al., Science 2014; Ye et al., Science 2014). For example, we found that common alleles of IRF7 tune the strength of an individual’s anti-viral response, and that genetic control of splicing is prevalent and important for the immune response (Ye et al., Genome Res 2018). Building on these studies, we developed systematic methods to analyze variants (Ray et al., Nat Comm 2021; Mouri, Nat Genetics, 2022). We also study non-genetic variations in human immunity, and found a myeloid cell type and state (‘MS1’ that corresponds to MDSCs) strongly associated with severe infections (bacterial and viral, including COVID-19) and sepsis (Reyes et al, Nat Med 2020, Science Tr Med 2021), leading us to new hypotheses underlying these dangerous clinical trajectories.
Drivers of autoimmunity
Deficiencies in nucleases that degrade DNA lead to accumulation of self DNA, activation of innate immune responses and development of autoimmune disorders, including systemic lupus erythematosus and Aicardi-Goutières syndrome in humans. How does autoimmunity develop upon triggering of innate immunity by self DNA (rather than pathogen-derived DNA)? We made the surprising observation that immunostimulatory DNA can arise from host damaged DNA that is exported from the nucleus to the lysosome (Lan et al., Cell Rep 2014). We hypothesize that this cellular process is a source of inflammation in autoimmunity, cancer, chemotherapy and aging. To further find drivers of autoimmunity, we’ve been analyzing kidney biopsies and blood from lupus patients in a small (Arazi et al., Nat Imm 2019) and large patient cohort (ongoing) and more recently in comparison to animal lupus models (Hoover et al., bioRxiv 2023).
Publications
Selected Publications
Chen JH, Nieman LT, Spurrell M, Jorgji V, Elmelech L, Richieri P, Xu KH, Madhu R, Parikh M, Zamora I, Mehta A, …Pelka K, Aryee MJ, Mino-Kenudson M, Gainor JF, Korsunsky I, Hacohen N. Human lung cancer harbors spatially organized stem-immunity hubs associated with response to immunotherapy. Nat Immunol. 2024. Apr;25(4):644-658
Liu B, Carlson RJ, Pires I, Gentili M, Feng E, Hellier Q, Schwartz MA, Blainey PC, Irvine DJ, Hacohen N. Human STING is a proton channel. Science 2023. Aug 4 ;381(6657):508-514.
Pelka K, Hofree M, Chen JH, Sarkizova S, Pirl JD, Jorgji V… Ng K, Giannakis M, Nieman LT, Boland GM, Aguirre AJ, Anderson AC, Rozenblatt-Rosen O, Regev A, Hacohen N. Spatially organized multicellular immune hubs in human colorectal cancer. Cell. 2021 Aug 24:S0092-8674(21)00945-4.
Reyes M, Filbin MR, Bhattacharyya RP, Sonny A, Mehta A, Billman K… Baron RM, Goldberg MB, Blainey PC, Hacohen N. Plasma from patients with bacterial sepsis or severe COVID-19 induces suppressive myeloid cell production from hematopoietic progenitors in vitro. Sci Transl Med. 2021 Jun 16;13(598):eabe9599.
Sarkizova S, Klaeger S, Le PM, Li LW, Oliveira G, Keshishian H, Hartigan CR, Zhang W, Braun DA, Ligon KL, Bachireddy P, Zervantonakis IK, Rosenbluth JM, Ouspenskaia T, Law T, Justesen S, Stevens J, Lane WJ, Eisenhaure T, Lan Zhang G, Clauser KR, Hacohen N, Carr SA, Wu CJ, Keskin DB. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat Biotechnol. 2020 Feb;38(2):199-209. doi: 10.1038/s41587-019-0322-9.
Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, Freeman SS, Reuben A, Hoover PJ, Villani A-C, Ivanova E, Portell A, Lizotte PH, Aref AR, Eliane JP, Hammond MR, Vitzthum H, Blackmon SM, Li B, Gopalakrishnan V, Reddy SM, Cooper ZA, Paweletz CP, Barbie DA, Stemmer- Rachamimov S, Flaherty KT, Wargo JA, Boland GM, Sullivan RJ, Getz G and Hacohen N. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018 Nov 1;175(4):998-1013.e20.
Ott P, Hu X, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, Chen C, Olive O, Carter TA, Li S, Lieb DJ, Eisenhaure T, Gjini E, Stevens J, Lane WJ, Javeri I, Nellaiappan K, Andreas Salazar12, Daley H, Seaman M, Buchbinder EI, Yoo CH, Harden M, Lennon N, Gabriel S, Rodig SJ, Barouch DH, Aster JC, Getz G, Wucherpfennig K, Neuberg D, Ritz J, Lander ES, Fritsch EF, Hacohen N & Wu CJ. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, Jul 13;547(7662):217-221.
*Equal contribution