Krantz Family Center for Cancer Research
Hwang Lab


Contact Information
William L. Hwang, MD, PhD
Assistant Professor of Radiation Oncology
Harvard Medical School
Associate Director, Radiation Biology & Research Program
Mass General Brigham Cancer Institute
Program Affiliations
Krantz Family Center for Cancer Research
Explore the Hwang Lab
2024 Krantz Awards Recipient
2024 Quantum Award: Boosting immunotherapy response while eradicating toxicity
Team: Lloyd Bod, PhD, William L. Hwang, MD, PhD, David T. Ting, MD, and Alexandra-Chloe Villani, PhD.
Learn about the team's project and the Krantz Awards
Research Summary
The Hwang laboratory focuses on the immense phenotypic, temporal and spatial heterogeneity of tumor ecosystems and the many insights that can only be gleaned by studying these systems at the level of their individual components. We study tumor-stroma interactions at unprecedented resolution through the development and application of techniques in spatial and systems oncology, advanced microscopy, genetic engineering and computational biology to patient-derived specimens, stromal tumoroids and mouse models. Our goals are to elucidate mechanisms of (1) therapeutic resistance mediated by genetic, epigenetic, and phenotypic factors including cell state plasticity; (2) treatment-mediated remodeling of the spatial microarchitecture of tumors and underlying cancer cell-stromal interactions; and (3) tumor-nerve crosstalk, which plays a critical role in the pathophysiology and morbidity of many malignancies but remains understudied.
Research Projects
Single-cell dynamics
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal and treatment refractory disease. Molecular subtyping of PDAC is rudimentary and does not currently inform clinical management or therapeutic development. We optimized single-nucleus RNA-seq to discover treatment-associated changes in cellular composition and state, including enrichment of a novel neurallike malignant program in residual tumors after chemoradiation. Our high-resolution molecular framework elucidates the inter- and intra-tumoral diversity of PDAC, treatment-associated remodeling and clinically relevant prognostication to enable precision oncology in PDAC.
Ongoing Projects:
- Identifying key regulators, context dependence and therapeutic vulnerabilities of resistant cell states
- Elucidating (epi)genetic contributions to cell state plasticity in therapeutic resistance
- Investigating mechanisms of tumorigenesis using single-cell multiomics to enable chemoprevention and early detection
- Studying developmental lineages and mechanisms of metastasis in pancreatic neuroendocrine tumors
Spatial oncology
Dissociative single-cell approaches enable detailed characterization of the different cell types and states that compose a heterogeneous tumor but sacrifice in situ spatial relationships among cells. Leveraging recent advances in spatial proteo-transcriptomics enabling single- cell resolution and high molecular plex, we performed spatial molecular profiling (SMI) on a cohort of patient-derived PDAC tumors and developed a novel method for inferring multicellular interactions. Spatially Constrained Optimal Transport Interaction Analysis (SCOTIA) that considers both spatial distance and ligand-receptor (LR) expression (collaborator: Martin Hemberg). We used SCOTIA to dissect the remodeled pancreatic tumor microenvironment in response to neoadjuvant chemoradiation and uncovered marked changes in LR interactions between cancer-associated f ibroblasts and malignant cells, which was supported by orthogonal experiments using a murine tumoroid co-culture system (https://tinyurl.com/2xtdytxt).
Overall, we demonstrated the immense potential of a translational spatial biology paradigm for deriving novel biological insights and identifying actionable therapeutic targets — one that can be broadly applied to other malignancies and treatment contexts.
Ongoing Projects:
- Identifying cell-type specific mediators of nerve outgrowth, invasion and colonization using patient-derived tumors, tumoroids and GEMMs
- Determining influence of neuronal subtype and activity on the immune response to cancer in primary tumors and draining lymph nodes
- Dissecting molecular mechanisms of dynamic physical interactions between cancer cells and nerves
- Discovering the mechanistic basis for differential central nervous system versus peripheral nervous system tropism across the spectrum of cancer
Publications
Selected Publications
Shiau C*, Cai J*, Gregory MT, Gong D, Yin X, Cho J-W, … Fernandez-del Castillo C, Mino-Kenudson M, Ting DT, Hemberg M†, Hwang WL†. Spatially resolved analysis of pancreatic cancer identifies therapyassociated remodeling of the tumor microenvironment. Nature Genetics 2024, online ahead of print.
Hwang WL*, Jagadeesh KA*, Guo JA*, Hoffman HI*, Yadollahpour P, Reeves J, … Fernandez-del Castillo C, Liss AS, Ting DT, Jacks T‡, Regev A‡. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nature Genetics 2022 Aug;54(8):1178-1191.
Shi DD, Guo JA, Hoffman HI, Su J, Mino-Kenudson M, Barth JL, Schenkel JM, Loeffler JS, Shih HA, Hong TS, Wo JY, Aguirre AJ, Jacks TJ, Zheng L, Wen PY, Wang TC, Hwang WL‡. Therapeutic avenues for cancer neuroscience: translational frontiers and clinical opportunities. Lancet Oncology. 2022;23(2):e62-74.
Guo, JA, Hoffman, HI, Weekes, CD, Zheng, LZ, Ting, DT, Hwang, WL‡. Refining the molecular framework for pancreatic cancer with single-cell and spatial technologies. Clinical Cancer Research. 2021;27(14):3825-3833.
Guo JA, Hoffman HI, Shroff S, Chen P, Hwang PG, Kim DY, Kim DW, Cheng SW, Zhao D, Mahal BA, Alshalafa M, Niemierko A, Wo JY, Loeffler JS, Fernandez-del Castillo C, Jacks T, Aguirre AJ, Hong TS, Mino-Kenudson M, Hwang WL‡. Pan-cancer transcriptomic predictors of perineural invasion improve occult histopathological detection. Clinical Cancer Research. 2021;27(10):2807-2815.
Hwang WL*, Pike LRG*, Royce TJ, Mahal BA, Loeffler JS‡. Safety of combining radiotherapy with immune checkpoint inhibitors. Nature Reviews Clinical Oncology. 2018;15(8):477-94.
Hwang WL*, Deindl S*, Harada BT, Zhuang X‡. Histone H4 tail mediates allosteric regulation of nucleosome remodeling by linker DNA. Nature. 2014;512(7513):213-7.
‡ Corresponding author
*Equal contribution
Research Image
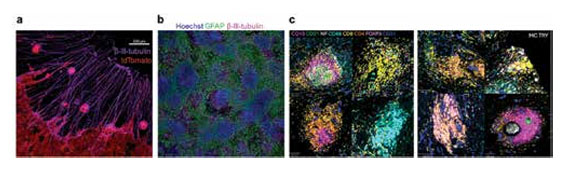
(a) Pancreatic cancer cell aggregates (red) moving along neurites (purple) extending from a dorsal root ganglion (removed). (b) Embryonic murine neurons in culture without glial depletion. (c) Representative immune aggregates in the pancreatic cancer tumor microenvironment identified by 100+ plex spatial proteomics. NF = neurofilament; MC TRY = mast cell tryptase.
Our Researchers
-
![]()
- Department of Radiation Oncology
Group Members
- Jake Adelman, BS†
- Beatrice Awasthi, PhD#
- Jung Woo Bae, MS^
- Leou Ismael Banla, MD, PhD#
- Hongyong Choi, MD, PhD##
- Hannah Donner, BS**
- Dennis Gong, BS*
- Dylan Harwood, BS†
- Nicole Lester, BS^
- Phuong Nguyen, PhD#
- Deniz Olgun†
- Ella Perrault, BS*
- Jeanna M. Qiu, BS**
- Daniel Rosen, MD, PhD#
- Peter L. Wang, PhD#
- Elizabeth (Xunqin) Yin, MD, PhD^
* Graduate student **
MD/PhD student (including rotation)
# Postdoctoral fellow/research scientist
## Visiting Professor
† Undergraduate student/visiting student
^ Staff
Krantz Family Center for Cancer Research
The scientific engine for discovery for the Mass General Brigham Cancer Institute.
Support the Krantz Family Center for Cancer Research
When you support us you are enabling discoveries that will lead to effective new weapons in the battle against cancer.

