Krantz Family Center for Cancer Research
Iafrate Lab


Contact Information
Iafrate Lab
Anthony John Iafrate, MD, PhD
Austin L. Vickery, Jr. Professor of Pathology
Deputy Chair, Department of Pathology
Program Affiliations
Krantz Family Center for Cancer Research
Department of Pathology Center for Integrated Diagnostics
Explore the Iafrate Lab
2024 Krantz Awards Recipient
2024 Spark Award: Comprehensive and cost-effective WGS-cfDNA assay: Breakthrough in ability to develop clinical genomic biomarkers and discover resistance mechanisms
Team: Gad A. Getz, PhD, Doğa Gülhan, PhD, Anthony John Iafrate, MD, PhD.
Research Summary
The Iafrate laboratory has focused efforts on developing highly complex molecular analyses of tumor genetics using novel technologies. We have a strong interest in the clinical implementation of genetic screening technologies that can help direct targeted therapies, focusing on lung, breast and brain tumors. Our recent contributions in the treatment of a subset of non-small cell lung carcinoma (NSCLC) with rearrangements of the ALK tyrosine kinase, rearrangements of the ROS1 tyrosine kinase and MET exon 14 skipping with a small molecule kinase inhibitor (crizotinib), underscore the promise of personalized cancer care (1, 2). We currently are focusing on detecting tumor DNA in blood samples (“liquid biopsies”) to allow for efficient and convenient tracking of cancer progression. In additional we are developing new techniques to allow for early detection of cancers by detecting tumor-specific DNA in circulation.
Research Projects
We have developed and deployed next generation sequencing to detect chromosomal rearrangements in tumor tissue, with on-going studies that assess the relative sensitivity in much larger clinical cohorts. The method we have developed, termed “anchored multiplex PCR” or AMP, is an efficient target enrichment technology, allowing for 100s of targets to be simultaneously analyzed from small tissue samples (3). We have used AMP to screen thousands of tumor samples, and have uncovered numerous novel driver fusion genes. Our lab is now focused on modeling novel fusions in vitro and developing therapeutic approaches to screening these fusions. We have also initiated studies of tumor heterogeneity; these efforts focus on gene amplification of receptor tyrosine kinases in glioblastoma (4). This work has revealed a new subclass of brain tumors with mosaic gene amplification of up to three kinases in distinct but intermingled cell populations within the same tumor, forming a mosaic pattern. We found that each subpopulation was actively proliferating and contributing to tumor growth. Detailed genetic analysis found that different subpopulations within a particular tumor shared other gene mutations, indicating that they had originated from the same precursor cells. Mapping the location of different subpopulations in the brain of a glioblastoma patient suggested that each subpopulation may serve a different function in the growth and spread of the tumor. Our lab has developed novel highly-multiplexed FISH technology to address how many genes show copy number heterogeneity, and to study the spatial distribution of such populations (5), see research image. We are exploring the therapeutic implications of such driver gene heterogeneity in cell line model systems of glioblastoma using genome-wide CRISPR knock out screens.
More recently we have adapted the AMP sequencing technology in other areas, including:- mapping off-target rates for CRISPR-CAS genome editing
- sequencing and mapping the distribution of IgH and TCR rearrangements in tumor samples
- ultra-high sensitive mutation calling in circulating tumor cells and cell free plasma samples
The lab has developed multiplex immunofluo rescence panels to study the spatial biology of tumor types including ovarian cancer (looking at homologous recombination repair proteins) and head and neck cancan (looking at immune infiltrates). Using the Lunaphore platform, the lab can simultaneously examine >15 markers at true singles cell resolution. We have development computation pipelines to analyze these complex datasets.
Publications
Selected Publications
Garcia-Beltran WF, lab EC, Astudillo MG, Yang D, Miller TE, Feldman J, Hauser BM, Caradonna TM, Clayton KL, Nitido AD, Murali MR, Alter G, Charles RC, Dighe A, Branda Ja, Lennerz JK, Lingwood D, Schmidt AG, Iafrate AJ, Balazs AB. Covid-19-neutralizing antibodies predict disease severity and survival. Cell. 2021; 21;184(2):476-488.
Cheng J, Cao Y, MacLeay A, Lennerz JK, Baig A, Frazier RP, Lee J, Hu K, Pacula M, Meneses E, Robinson H, Batten JM, Brastianos PK, Heist RS, Bardia A, Le LP, Iafrate AJ. Clinical Validation of a Cell-Free DNA Gene Panel. J Mol Diagn. 2019; 21(4): 632-645.
Onozato ML, Yapp C, Richardson D, Sundaresan T, Chahal V, Lee J, Sullivan JP, Madden MW, Shim HS, Liebers M, Ho Q, Maheswaran S, Haber DA, Zheng Z, Clancy B, Elliott HL, Lennerz JK, Iafrate AJ. Highly Multiplexed Fluorescence in Situ Hybridization for in Situ Genomics. J Mol Diagn. 2019; 21(3):390-407.
Heist RS, Shim HS, Gingipally S, Mino- Kenudson M, Le L, Gainor JF, Zheng Z, Aryee M, Xia J, Jia P, Jin H, Zhao Z, Pao W, Engelman JA, and Iafrate AJ. MET Exon 14 Skipping in Non-Small Cell Lung Cancer. Oncologist. 2016; 21(4):481-486.
Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Chen J, Robinson HE, Chmielecki J, Pao W, Engelman JA, Iafrate AJ*, Le LP*. Anchored multiplex PCR for targeted nextgeneration sequencing. Nat Medicine. 2014; Nov. 10.
Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon B, Salgia R, Riely GJ, Varella-Garcia M, Shapiro GI, Costa DB, Doebele RC, Le LP, Zheng Z, Tan W, Stephenson P, Shreeve SM, Tye LM, Christensen JG, Wilner K, Clark JW, Iafrate AJ. Crizotinib in ROS1-Rearranged Non-Small Cell Lung Cancer. N Engl J Med. 2014; Sept. 27.
*Co-corresponding authors
Research Image
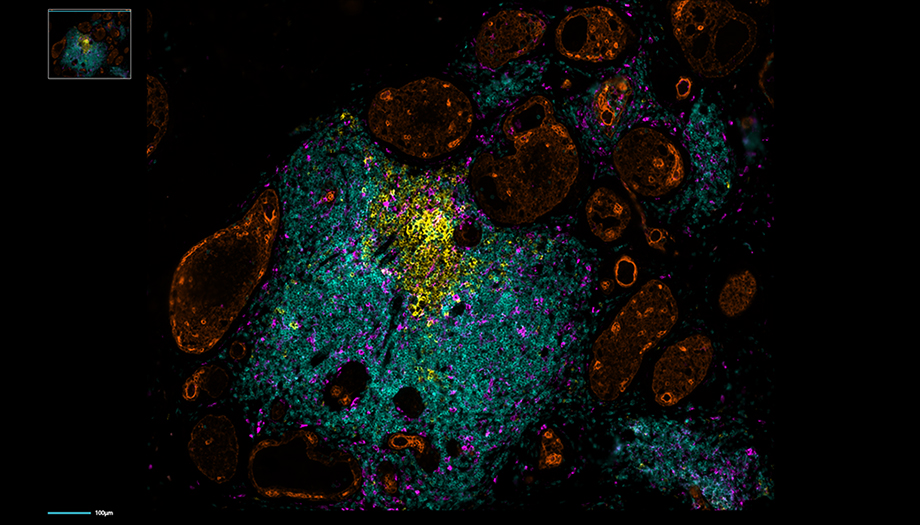
Multiplex Immunofluorescence to detect changes in the immune landscape in head and neck tumors.
Our Researchers
-
![]()
- Austin L. Vickery, Jr. Professor of Pathology
- Vice Chair of Pathology for Academic Affairs
- Pathologist in Pathology, Massachusetts General Hospital
Lab Members
- Kellie A. Burke†
- Elizabeth Codd
- Samantha Flynn
- JoAnne Fordman, PhD
- Anna Gao, PhD
- Helen Hou
- Stefan Kaluziak, PhD
- Prinjali Kalyan
- Kevin Kim*
- Annie Li, MD
- Ka Yee Li, PhD
- Dawn Mitchell*
- Cam Nguyen
- Natalie Nordenfelt
- Mihir Shrestha
- Sandesh Timilsina
- Diane Yang, PhD
- Edwin Zhang *
* Graduate student
† Admin Assistant
Krantz Family Center for Cancer Research
The scientific engine for discovery for the Mass General Brigham Cancer Institute.
Support the Krantz Family Center for Cancer Research
When you support us you are enabling discoveries that will lead to effective new weapons in the battle against cancer.

